Featured publications
For a full list of publications see PubMed or here.
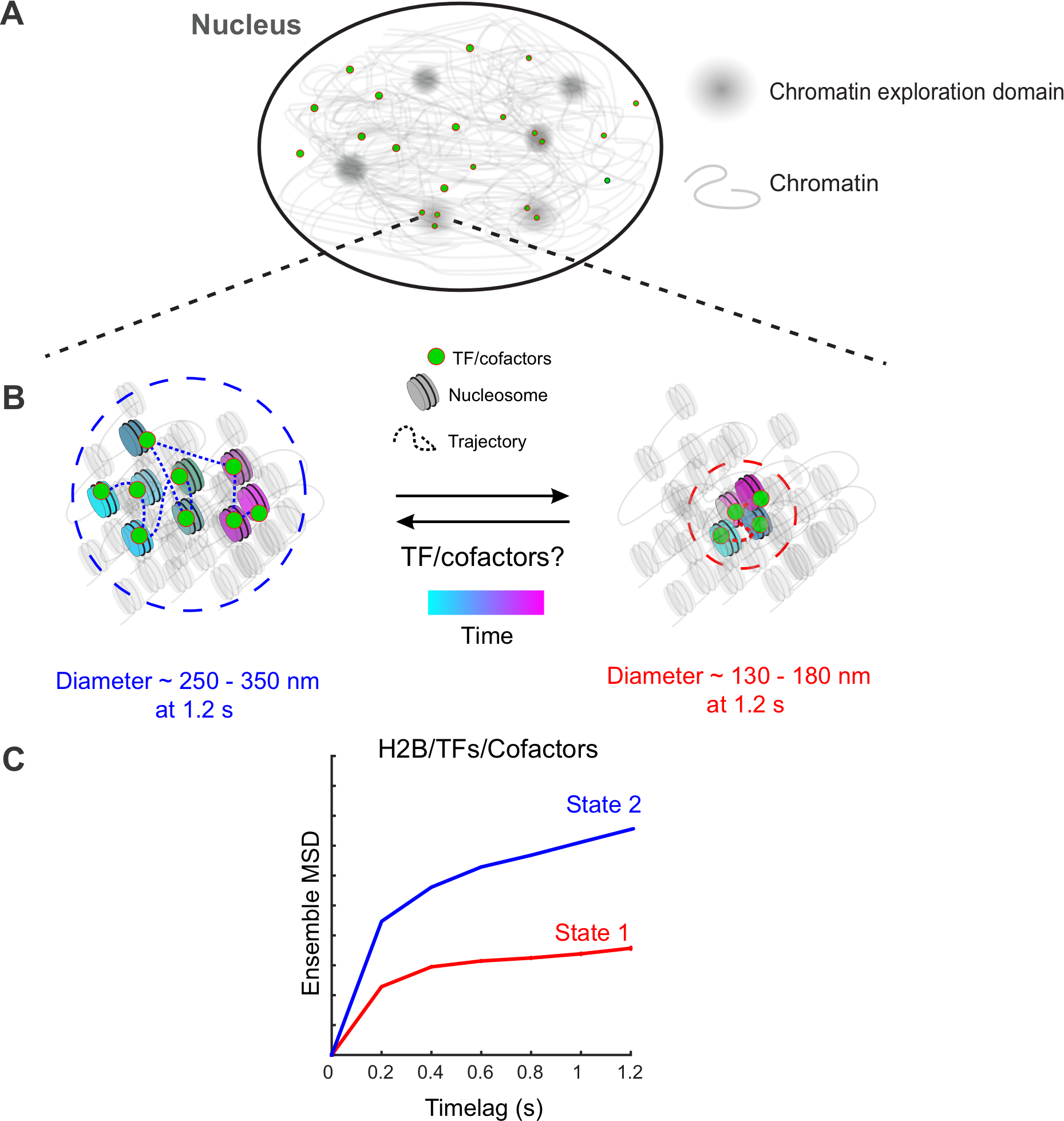
Science Advances 2023
Wagh KS, Stavreva DA, Jensen RAM, Paakinaho V, Fettweis G, Schiltz RL, Wustner D, Mandrup S , Presman DM, Upadhyaya A and Hager GL
How chromatin dynamics relate to transcriptional activity remains poorly understood. Using single-molecule tracking, coupled with machine learning, we show that histone H2B and multiple chromatin-bound transcriptional regulators display two distinct low-mobility states. Ligand activation results in a marked increase in the propensity of steroid receptors to bind in the lowest-mobility state. Mutational analysis revealed that interactions with chromatin in the lowest-mobility state require an intact DNA binding domain and oligomerization domains. These states are not spatially separated as previously believed, but individual H2B and bound-TF molecules can dynamically switch between them on time scales of seconds. Single bound-TF molecules with different mobilities exhibit different dwell time distributions, suggesting that the mobility of TFs is intimately coupled with their binding dynamics. Together, our results identify two unique and distinct low-mobility states that appear to represent common pathways for transcription activation in mammalian cells.
[pubmed] [Sci. Adv.]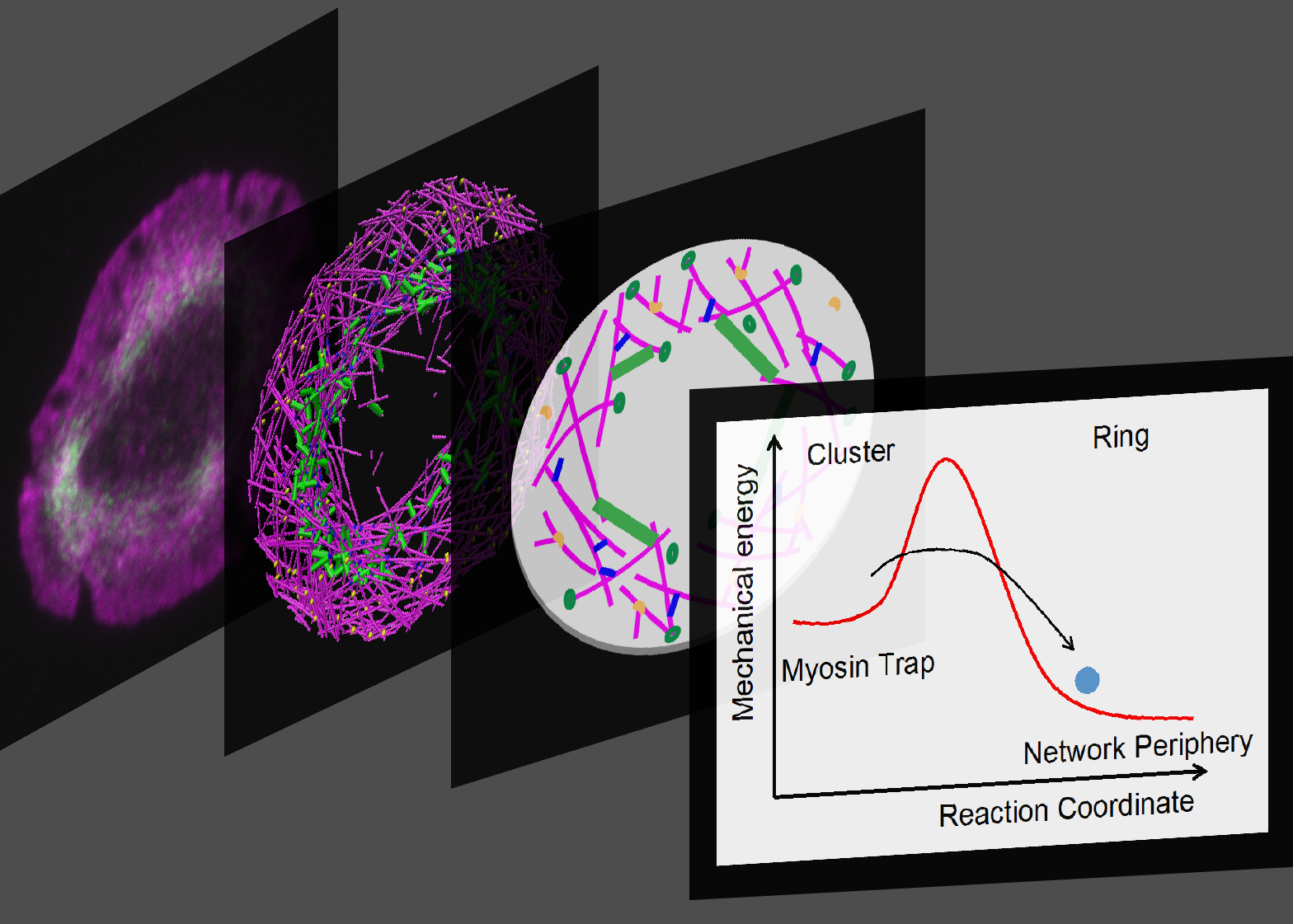
eLife 2022
Ni Q, Wagh K, Pathni A, Ni H, Vashisht V, Upadhyaya A and Papoian G
In most eukaryotic cells, actin filaments assemble into a shell-like actin cortex under the plasma membrane, controlling cellular morphology, mechanics, and signaling. The actin cortex is highly polymorphic, adopting diverse forms such as the ring-like structures found in podosomes, axonal rings, and immune synapses. The biophysical principles that underlie the formation of actin rings and cortices remain unknown. Using a molecular simulation platform, called MEDYAN, we discovered that varying the filament treadmilling rate and myosin concentration induces a finite size phase transition in actomyosin network structures. We found that actomyosin networks condense into clusters at low treadmilling rates or high myosin concentration but form ring-like or cortex-like structures at high treadmilling rates and low myosin concentration. This mechanism is supported by our corroborating experiments on live T cells, which exhibit ring-like actin networks upon activation by stimulatory antibody. Upon disruption of filament treadmilling or enhancement of myosin activity, the pre-existing actin rings are disrupted into actin clusters or collapse towards the network center respectively. Our analyses suggest that the ring-like actin structure is a preferred state of low mechanical energy, which is, importantly, only reachable at sufficiently high treadmilling rates.
[eLife]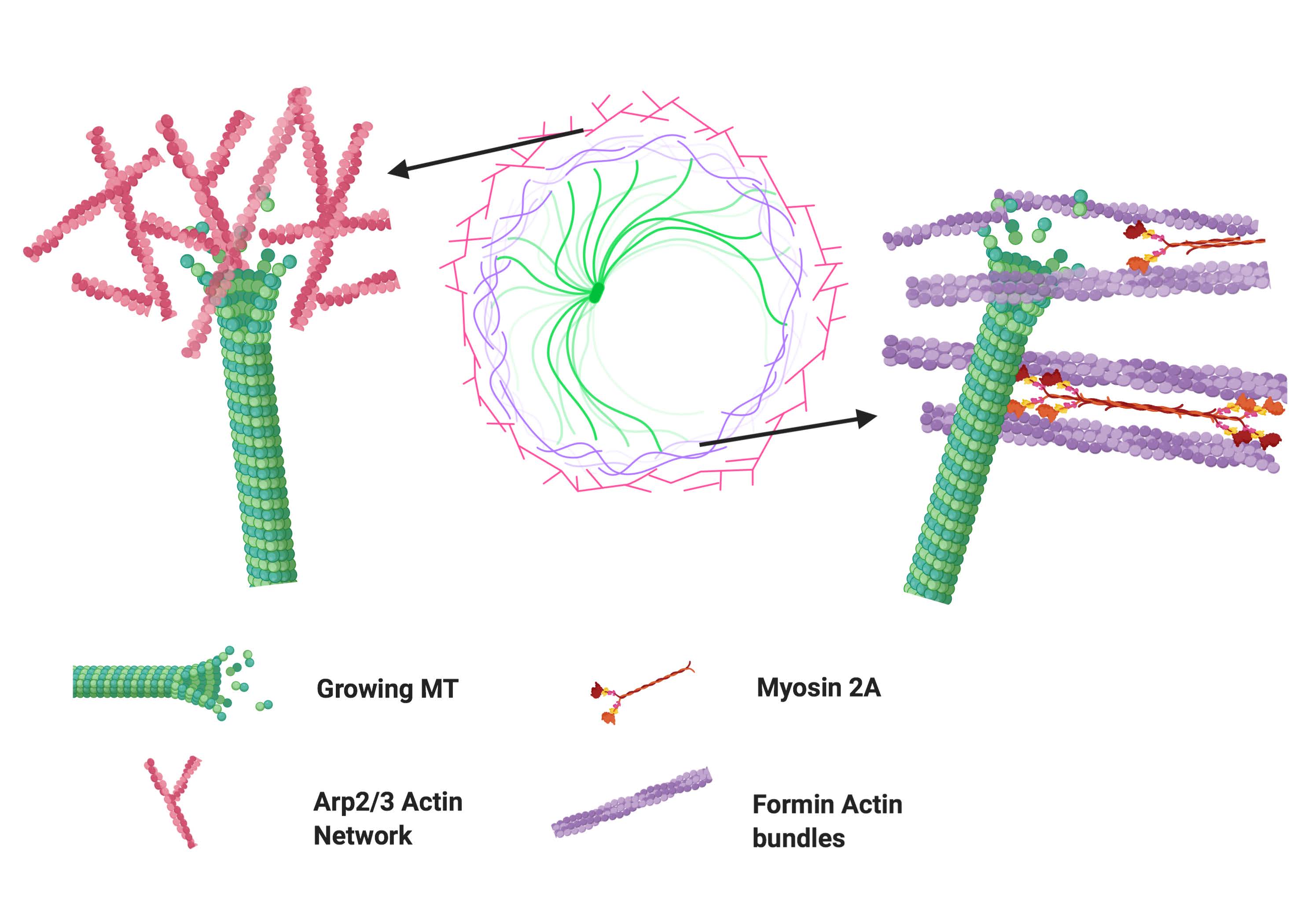
Molecular Biology of the Cell 2021
Rey-Suarez I, Rogers N, Kerr S, Shroff H, and Upadhyaya A
Activation of T cells leads to the formation of the immunological synapse (IS) with antigen presenting cells. This requires T cell polarization and coordination between the actomyosin and microtubule cytoskeletons. The interactions between these two cytoskeletal components during T cell activation are not well understood. Here, we elucidate the interactions between microtubules and actin at the IS with high-resolution fluorescence microscopy. We show that microtubule growth dynamics in the peripheral actin-rich region are distinct from those in the central actin-free region. We further demonstrate that these differences arise from differential involvement of Arp2/3- and formin-nucleated actin structures. Formin inhibition results in a moderate decrease in microtubule growth rates, which is amplified in the presence of integrin engagement. In contrast, Arp2/3 inhibition leads to an increase in microtubule growth rates. We find that microtubule filaments are more deformed and exhibit greater shape fluctuations in the periphery of the IS compared to the center. Using small molecule inhibitors, we show that actin dynamics and actomyosin contractility play key roles in defining microtubule deformations and shape fluctuations. Our results indicate a mechanical coupling between the actomyosin and microtubule systems during T cell activation, whereby different actin structures influence microtubule dynamics in distinct ways.
[pubmed] [MBoC]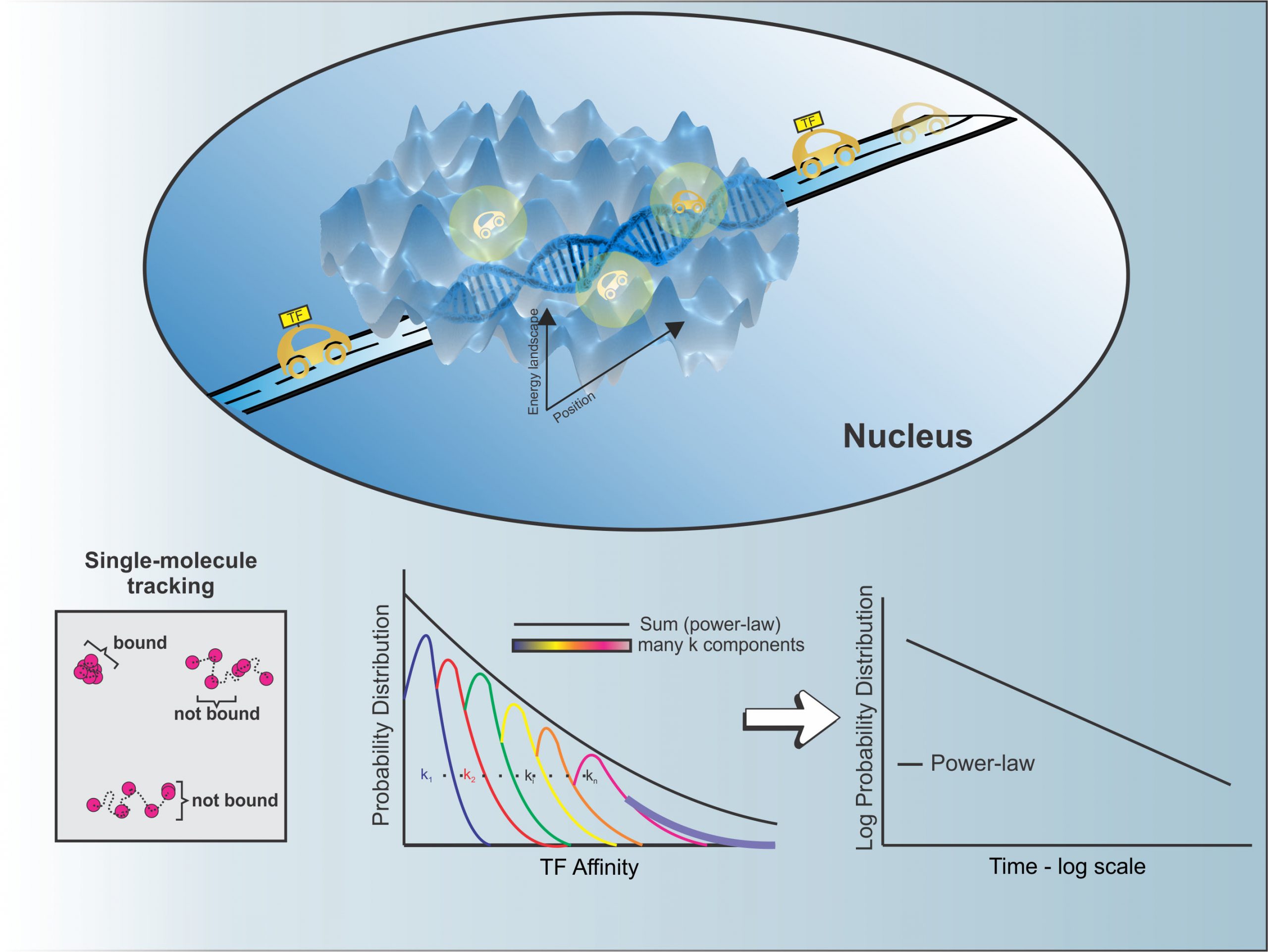
Nucleic Acids Research 2021
Garica DA, Fettweis GJ, Presman DM, Paakinaho V, Jarzynski C, Upadhyaya A, and Hager GL
Single-molecule tracking (SMT) allows the study of transcription factor (TF) dynamics in the nucleus, giving important information regarding the diffusion and binding behavior of these proteins in the nuclear environment. Dwell time distributions obtained by SMT for most TFs appear to follow bi-exponential behavior. This has been ascribed to two discrete populations of TFs—one non-specifically bound to chromatin and another specifically bound to target sites, as implied by decades of biochemical studies. However, emerging studies suggest alternate models for dwell-time distributions, indicating the existence of more than two populations of TFs (multi-exponential distribution), or even the absence of discrete states altogether (power-law distribution). Here, we present an analytical pipeline to evaluate which model best explains SMT data. We find that a broad spectrum of TFs (including glucocorticoid receptor, oestrogen receptor, FOXA1, CTCF) follow a power-law distribution of dwell-times, blurring the temporal line between non-specific and specific binding, suggesting that productive binding may involve longer binding events than previously believed. From these observations, we propose a continuum of affinities model to explain TF dynamics, that is consistent with complex interactions of TFs with multiple nuclear domains as well as binding and searching on the chromatin template.
[pdf] [Nucleic Acids Research]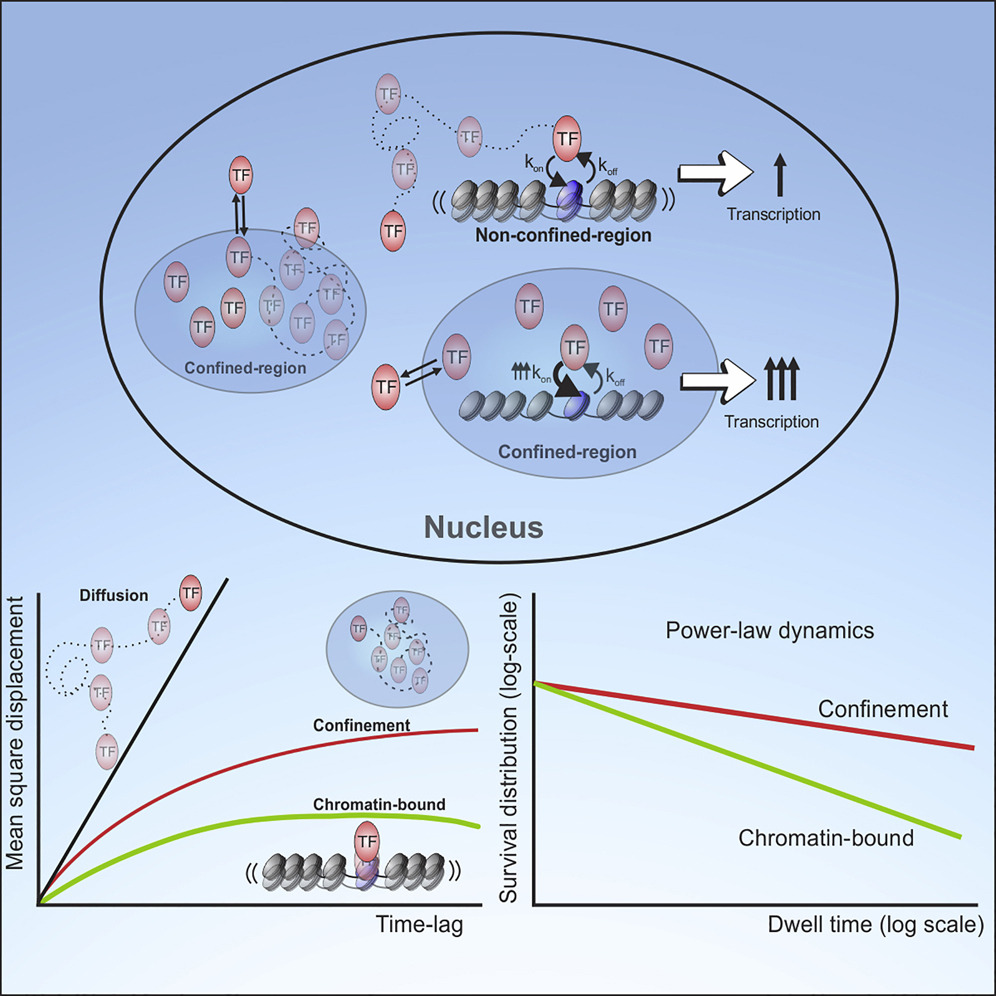
Molecular Cell 2021
Garica DA, Johnson TA, Presman DM, Fettweis G, Wagh KS, Rinaldo, L, Stavreva DA, Paakinaho V, Jensen RAM, Mandrup S, Upadhyaya A, and Hager GL
Transcription factors (TFs) regulate gene expression by binding to specific consensus motifs within the local chromatin context. The mechanisms by which TFs navigate the nuclear environment as they search for binding sites remain unclear. Here, we used single-molecule tracking and machine-learning-based classification to directly measure the nuclear mobility of the glucocorticoid receptor (GR) in live cells. We revealed two distinct and dynamic low-mobility populations. One accounts for specific binding to chromatin, while the other represents a confinement state that requires an intrinsically disordered region (IDR), implicated in liquid-liquid condensate subdomains. Further analysis showed that the dwell times of both subpopulations follow a power-law distribution, consistent with a broad distribution of affinities on the GR cistrome and interactome. Together, our data link IDRs with a confinement state that is functionally distinct from specific chromatin binding and modulates the transcriptional output by increasing the local concentration of TFs at specific sites.
[pdf] [Molecular Cell]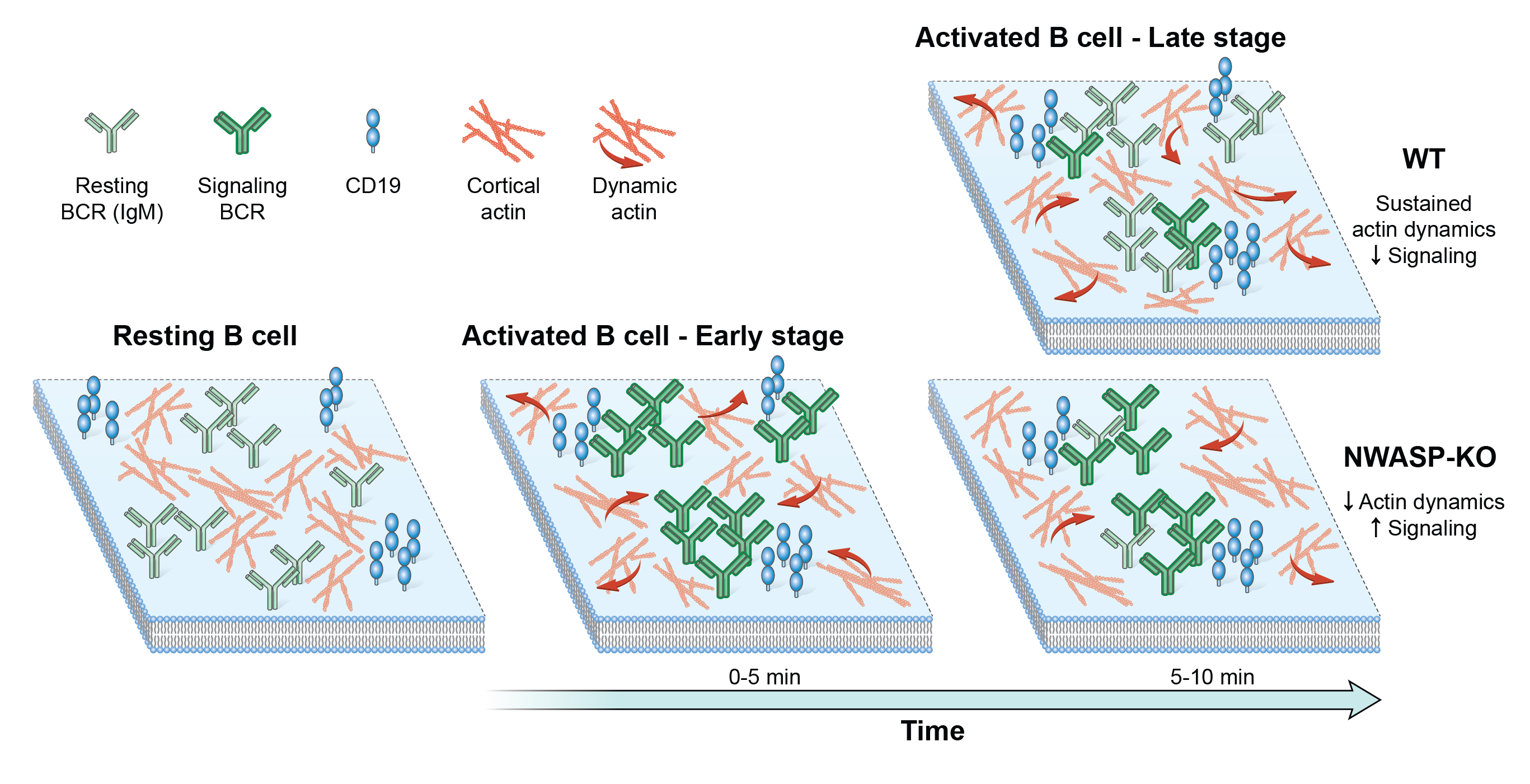
Nat. Commun. 2020
Rey-Suarez I, Wheatley BA, Koo P, Bhanja A, Shu Z, Mochrie S, Song W, Shroff H and Upadhyaya, A
Regulation of membrane receptor mobility tunes cellular response to external signals, such as in binding of B cell receptors (BCR) to antigen, which initiates signaling. However, whether BCR signaling is regulated by BCR mobility, and what factors mediate this regulation, are not well understood. Here we use single molecule imaging to examine BCR movement during signaling activation and a novel machine learning method to classify BCR trajectories into distinct diffusive states. Inhibition of actin dynamics downstream of the actin nucleating factors, Arp2/3 and formin, decreases BCR mobility. Constitutive loss or acute inhibition of the Arp2/3 regulator, N-WASP, which is associated with enhanced signaling, increases the proportion of BCR trajectories with lower diffusivity. Furthermore, loss of N-WASP reduces the diffusivity of CD19, a stimulatory co-receptor, but not that of FcγRIIB, an inhibitory co-receptor. Our results implicate a dynamic actin network in fine-tuning receptor mobility and receptor-ligand interactions for modulating B cell signaling.
[pdf] [Nat. Commun.]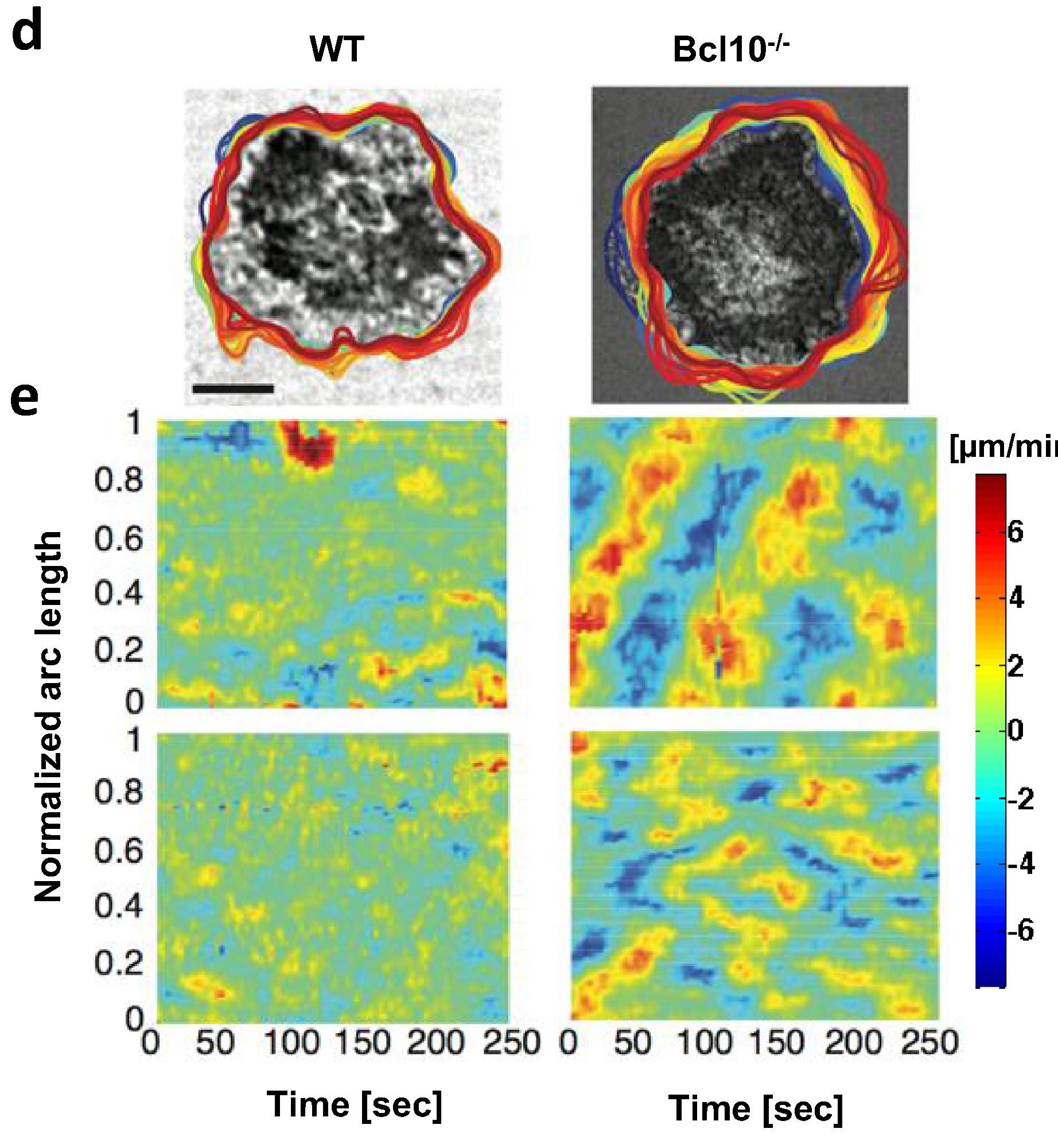
Cellular Immunology 2020
Wagh KS, Wheatley BA, Traver MK, Hussain I, Schaefer BC and Upadhyaya A
T cell responses to antigen are initiated by engagement of the T cell receptor (TCR)1, leading to activation of diverse signaling cascades, including an incompletely defined pathway that triggers rapid remodeling of the actin cytoskeleton. Defects in the control of actin dynamics and organization are associated with several human immunodeficiency diseases, emphasizing the importance of cytoskeletal remodeling in the functioning of the adaptive immune system. Here, we investigate the role of the adaptor protein Bcl102 in the control of actin dynamics. Although Bcl10 is primarily known as a component of the pathway connecting the TCR to activation of the NF-κB3 transcription factor, a few studies have implicated Bcl10 in antigen receptor-dependent control of actin polymerization and F-actin-dependent functional responses. However, the role of Bcl10 in the regulation of cytoskeletal dynamics remains largely undefined. To investigate the contribution of Bcl10 in the regulation of TCR-dependent cytoskeletal dynamics, we monitored actin dynamics at the immune synapse of primary murine CD8 effector T cells. Quantification of these dynamics reveals two distinct temporal phases distinguished by differences in speed and directionality. Our results indicate that effector CD8 T cells lacking Bcl10 display faster actin flows and more dynamic lamellipodia, compared to wild-type cells. These studies define a role for Bcl10 in TCR-dependent actin dynamics, emphasizing that Bcl10 has important cytoskeleton-directed functions that are likely independent of its role in transmission of NF-κB -activating signals.
[pubmed] [pdf] [Cellular Immunology]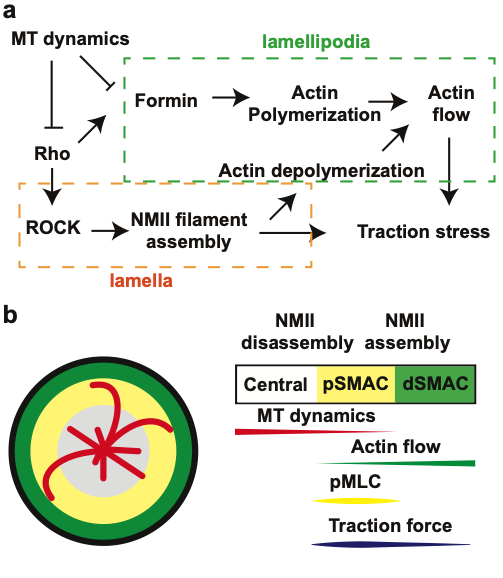
PNAS 2017
Hui KL and Upadhyaya A
T-cell receptor (TCR) triggering and subsequent T-cell activation are essential for the adaptive immune response. Recently, multiple lines of evidence have shown that force transduction across the TCR complex is involved during TCR triggering, and that the T cell might use its force-generation machinery to probe the mechanical properties of the opposing antigen-presenting cell, giving rise to different signaling and physiological responses. Mechanistically, actin polymerization and turnover have been shown to be essential for force generation by T cells, but how these actin dynamics are regulated spatiotemporally remains poorly understood. Here, we report that traction forces generated by T cells are regulated by dynamic microtubules (MTs) at the interface. These MTs suppress Rho activation, nonmuscle myosin II bipolar filament assembly, and actin retrograde flow at the T-cell-substrate interface. Our results suggest a novel role of the MT cytoskeleton in regulating force generation during T-cell activation.
[pdf] [PNAS]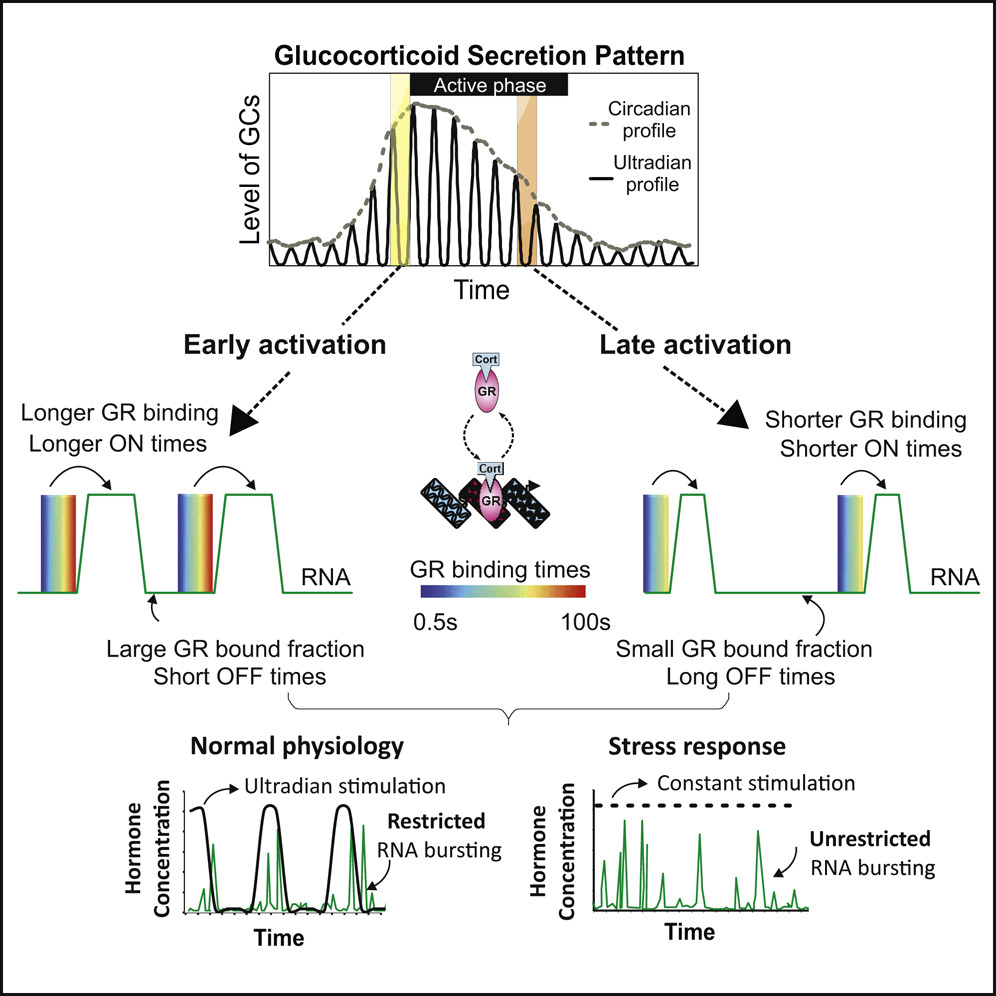
Molecular Cell 2019
Stavreva DA, Garcia DA, Fettweis G, Pegoraro G,… Upadhyaya A and Hager, GL
Genes are transcribed in a discontinuous pattern referred to as RNA bursting, but the mechanisms regulating this process are unclear. Although many physiological signals, including glucocorticoid hormones, are pulsatile, the effects of transient stimulation on bursting are unknown. Here we characterize RNA synthesis from single-copy glucocorticoid receptor (GR)-regulated transcription sites (TSs) under pulsed (ultradian) and constant hormone stimulation. In contrast to constant stimulation, pulsed stimulation induces restricted bursting centered around the hormonal pulse. Moreover, we demonstrate that transcription factor (TF) nuclear mobility determines burst duration, whereas its bound fraction determines burst frequency. Using 3D tracking of TSs, we directly correlate TF binding and RNA synthesis at a specific promoter. Finally, we uncover a striking co-bursting pattern between TSs located at proximal and distal positions in the nucleus. Together, our data reveal a dynamic interplay between TF mobility and RNA bursting that is responsive to stimuli strength, type, modality, and duration.
[pubmed] [pdf] [Molecular Cell]