Research in the Upadhyaya Lab
The research in our lab broadly pursues two themes: (1) How the physical properties of the cell and its environment influence biochemical signaling, cellular force generation and force response (with particular emphasis on the immune cells), and (2) How these biochemical and mechanical signals are relayed to the nucleus to regulate gene expression. In order to answer these questions, we use state of the art imaging for the visualization of internal protein dynamics at high spatial and temporal resolution, measurements of cellular forces simultaneously with signaling, single molecule and high-throughput genomics to measure gene expression, and genetic and physical tools to perturb the system.
Aashli Pathni, Vishavdeep Vashisht, Matthew Connell, Frank Fazekas, Mikayla Griener, Kiet Nguyen, Elizabeth Kiely
Funding: NIH MIRA
Cytoskeletal dynamics and signaling in T cells
T cell activation by antigen-presenting cells involves extensive changes in the actin and microtubule cytoskeletons. Cytoskeletal dynamics lead to the reorganization of receptors and signaling molecules into dynamic microclusters within the T cell/antigen-presenting cell interface. We use live cell and super-resolution imaging combined with quantitative image analysis and biophysical models to elucidate the molecular mechanisms that link T cell signaling and cytoskeletal dynamics. We have recently demonstrated that dynamic microtubules regulate actin assembly with the interface (Hui and Upadhyaya, 2017). We use super-resolution imaging methods (e.g. instant Structured Illumination Microscopy, in collaboration with Dr. Hari Shroff at NIBIB) and quantitative image analysis to uncover the molecular basis of the crosstalk between the actin and microtubule cytoskeletons during T cell activation (Rey-Suarez et al. 2021). We are also developing methods to optogenetically perturb T cell actin dynamics to dissect the signaling pathways that control cytoskeletal dynamics during this process. We work in collaboration with the Papoian group at UMD to develop active matter based models of cytoskeletal dynamics to gain a biophysical understanding of the origins and functional implications of these processes.
Ivan Rey-Suarez
Funding: NSF
Immune receptor dynamics
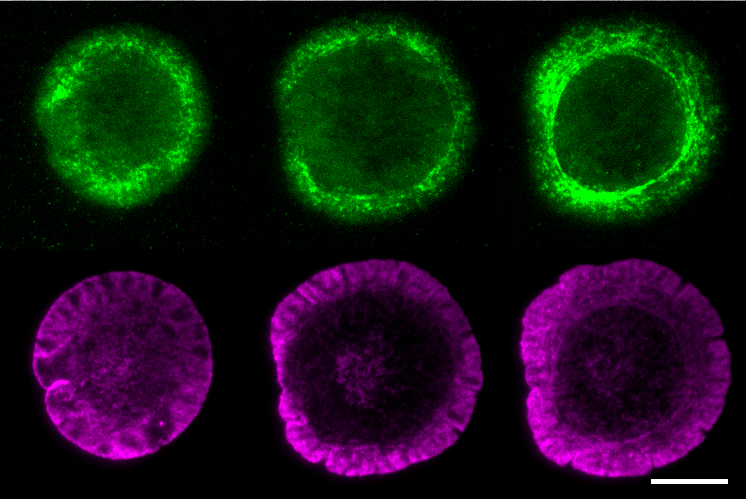
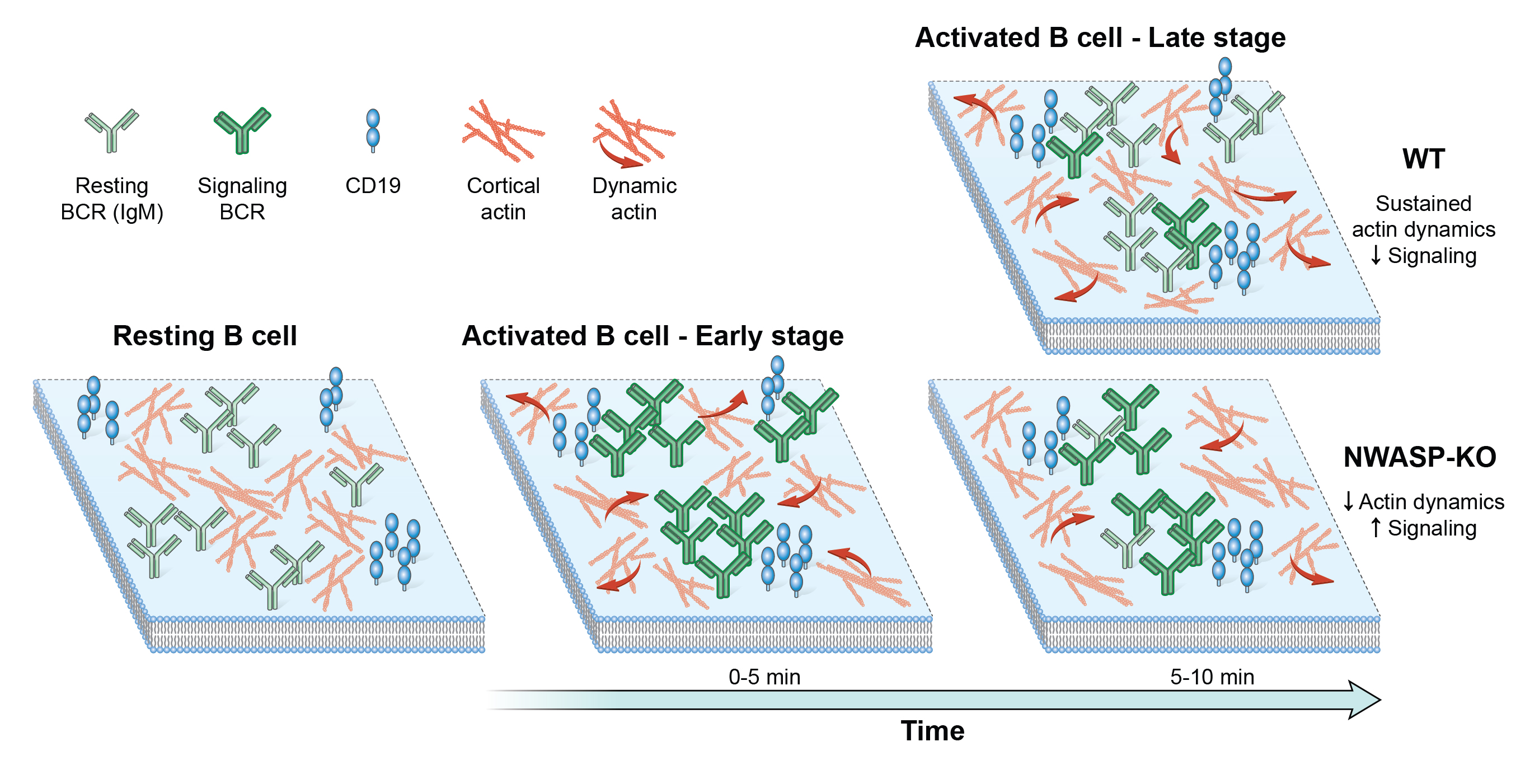
Cellular processes are often governed by molecular interactions or events, such as binding of cell surface receptors to extracellular ligand. These molecular scale events lead to cooperative behavior at the mesoscale such as aggregation into macromolecular complexes and emergence of larger scale cellular responses. How these different scales are linked is an open question. Mobility of single cell surface molecules can provide insight into their biochemical state, the physical properties of their microenvironment and the forces acting on them. Our work has shown that lateral mobility of ligands is an important parameter regulating microcluster formation and signaling threshold in B lymphocytes (Ketchum et al. 2014). A recent goal of our lab has been to dissect the biochemical interactions and evolution of signaling states. We have established imaging techniques to quantify movement of single B cell receptors with high spatial and temporal resolution. In collaboration with Simon Mochrie (Yale Univ.), we have used machine-learning methods to reveal the diffusive states populated by BCR and map these states onto BCR signaling states. We have shown that actin regulatory proteins (WASP and NWASP) modulate these diffusive and signaling states, indicating that actin based forces are important in driving receptor mobility, which in turn determines clustering and the biochemical response of B cells (Rey-Suarez et al. 2020). We are now developing analysis and imaging methods to study how active fluctuations drive B cell receptor movement and clustering.
Aashli Pathni, Mikayla Greiner, Elizabeth Kiely
Funding: NIH MIRA, NSF GRFP
Mechanosensing and force generation in T cells
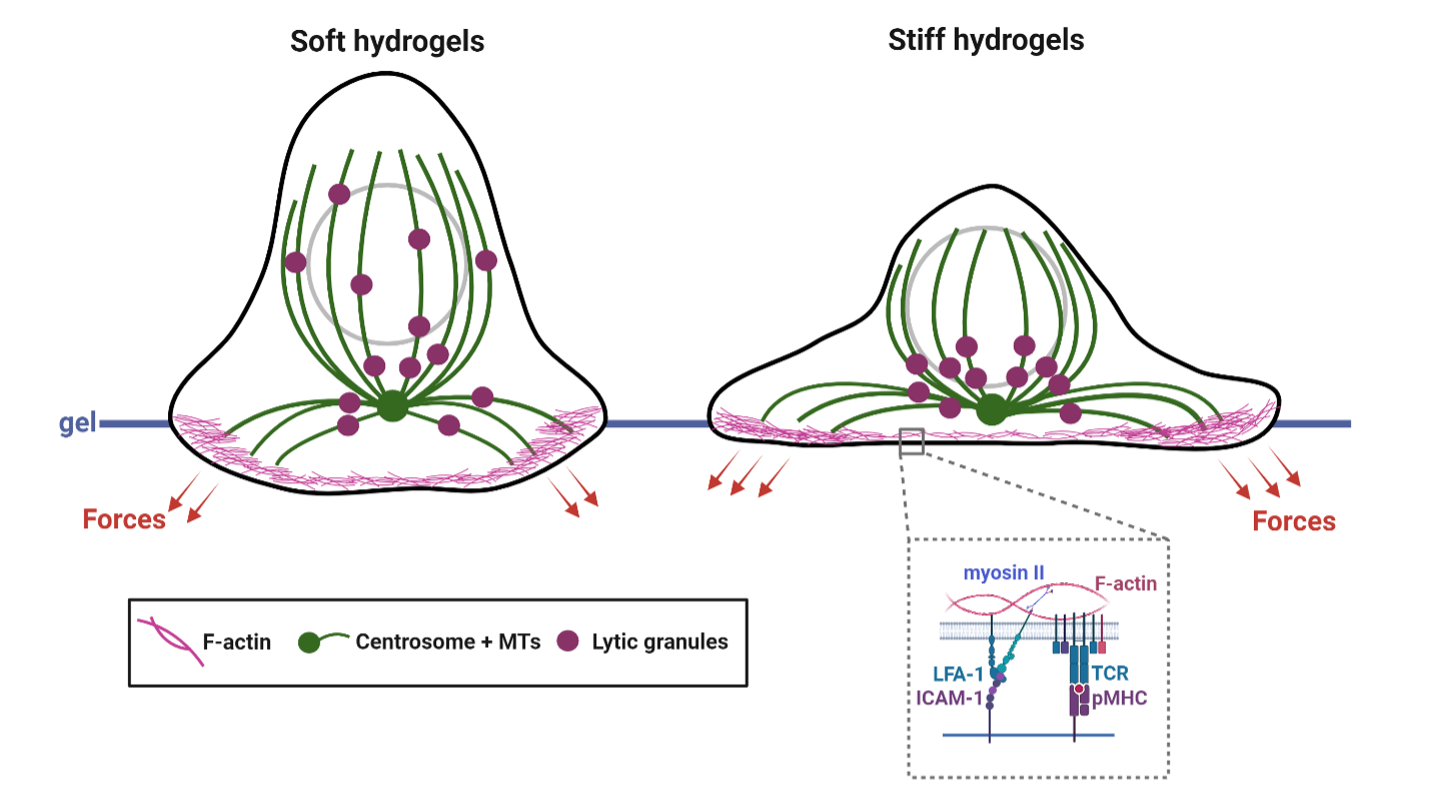
A number of studies have shown that signaling by single and ensembles of T/B cell receptors are sensitive to externally applied forces. However, in the body, these forces have to be generated by the cell itself. The magnitude, spatial organization and temporal properties of internal cytoskeletal forces and whether they are sufficient to engage signaling were not known. We have shown that while T cells are weak force generators, the forces are sufficient to facilitate receptor activation, and that the forces are largely generated and maintained by the dynamics of the acto-myosin cytoskeleton (Hui et al. 2014). These measurements demonstrate that physiological level of forces are sufficient for biochemical activation of signaling receptors. More recently we have shown a surprising and hitherto unknown role for microtubule filament dynamics in T cell force generation (Hui and Upadhyaya, 2017), and that the interaction between the actin and microtubule cytoskeletons is essential for optimum force balance in cells. Our ongoing work explores the biophysical mechanisms of force generation in T cells, its molecular regulation and functional impact. We are currently interested in understanding how cytokine signaling regulates cellular force generation in cytotoxic T cells and how this regulation affects their cytotoxic potential. Given the importance of dynamic force application in T cell activation, we are also interested in quantifying the statistics of force fluctuations during signaling and how they arise from active forcing by the cytoskeleton.

Vishavdeep Vashisht, Shankar Sivarajan, Matthew Connell
Funding: NSF Transitions, NIH MIRA
Mechanotransduction and gene expression
The molecular and biophysical mechanisms by which the physical environment influences cell fate and behavior are open questions in the field. Beyond the modulation of cellular processes such as cell shape, migration, signaling, cytoskeletal organization and dynamics (which has been our focus), the ultimate consequence of mechanosensing is the modulation of gene expression and changes in cell fate. Forces generated by the cytoskeleton as the cell adapts to mechanical stimuli are transmitted to the nucleus and chromatin via physical links on the nuclear envelope. How cytoskeletal forces affect nuclear dynamics and chromatin accessibility to transcription factors to drive gene expression are not well understood. Also, most insights about gene regulation by mechanical inputs have been gained from bulk analysis of gene expression patterns, which makes it difficult to link how forces generated by individual cells affect chromatin accessibility and transcription factor dynamics driving gene regulation (Wagh et al. 2021). We collaborate with Dr. Gordon Hager at NIH to use single molecule imaging of transcription factors as they diffuse and bind to cognate response elements on chromatin. We have recently demonstrated that transcription factor binding strengths are scale-free (Garica et al. 2021a) and intrinsic disordered regions on transcription factors alter their diffusion and transcriptional efficacy (Garcia et al. 2021b). We are currently studying how transcription factor dynamics and gene expression are altered by substrate stiffness.