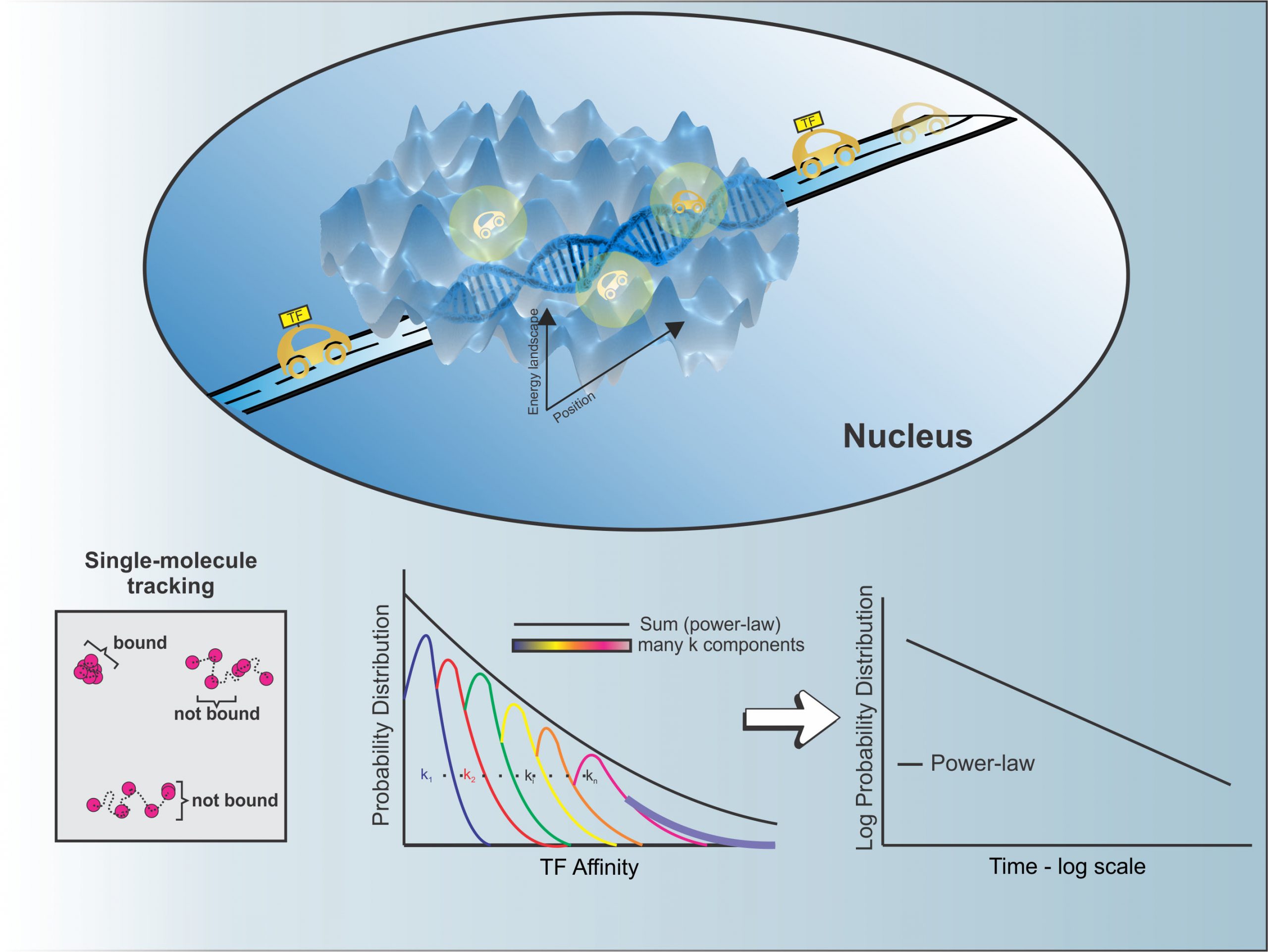
David’s paper on transcription factor dynamics is published as a “Breakthrough Article” in Nucleic Acids Research. What began as an improved photo-bleaching correction for single molecule dynamics in the nucleus evolved into an exciting new finding! Transcription factors (TF) must engage with chromatin to regulate gene expression. Single-molecule tracking makes it possible to follow individual TF molecules inside nuclei of live cells, where they diffuse and interact with chromatin. Current interpretation of the dwell-times, which are a measure of the interaction times with chromatin invoke a bi-exponential model, with a fast component representing nonspecific interactions with chromatin, while a slower component represents specific binding.
Using this new photobleaching correction, David showed that the bi-exponential model misrepresents the true interaction behavior of TFs. He, along with Diego, showed that transcription factor dwell time distributions are best described by a power-law model. A simple model, modified from the seminal work of Bouchaud and Georges, shows that the observed power-law distributions emerge as a result of a broad distribution of transcription factor-chromatin interaction affinities.